Data Standardization#
FAIR Principles Met#
FAIR PRINCIPLE I1
(Meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation
FAIR PRINCIPLE I2
(Meta)data use vocabularies that follow the FAIR principles
FAIR PRINCIPLE I3
(Meta)data include qualified references to other (meta)data
Introduction#
Data standardization is the process of transforming data into a common format or structure to ensure consistency, comparability, and compatibility across different datasets or systems. It involves cleaning, formatting, and organizing data to adhere to predefined norms or guidelines, making it easier to analyze, integrate, and share information effectively.
In the biodiversity domain the Biodiversity Information Standards (TDWG) is a non-profit organization and a community dedicated to developing biodiversity information standards. The TDWG is involved in developing, ratifying and promoting standards and guidelines for the recording and exchange of data about organisms; and acting as a forum for discussing all aspects of biodiversity information management through meetings, online discussions, and publications. A list of all standards in TDWG auspicious can be found at: https://www.tdwg.org/standards/.
Among the TDWG standards, the Darwin Core standard is widely used to document occurrence data.
It includes a glossary of terms (in other contexts these might be called properties, elements, fields, columns, attributes, or concepts) intended to facilitate the sharing of information about biological diversity by providing identifiers, labels, and definitions. Darwin Core is primarily based on taxa, their occurrence in nature as documented by observations, specimens, samples, and related information.
Despite it has been initially defined for occurrence data, it has been used for documeting other kinds of data, such as biotic interactions. Due its flexibility and capability to be extended it has been used within other vocabularies and ontologies.
Biotic interactions and the Darwin Core standard#
The Darwin Core (DwC) provides many different terms for documenting biotic interactions (e.g. associatedTaxa, associatedOccurrences, ResourceRelationship). However, we recommend the usage of the schema proposed in Salim et al. [SSZ+22], which is based on the Darwin Core Event and ResourceRelationship classes for representing biotic interactions. According to the authors the proposed schema:
The schema for biotic interactions has its grounds in the co-action definition proposed by Haskell [Has49], further refined by Lidicker [Lid79] for biotic interactions, and more recent, the biotic interactions model of interaction events introduced by Gómez et al. [GomezIT23].
Instances of DwC Event class serve as representations of interaction events. These instances capture essential information about the interactions, such as temporal data and sampling details. Additionally, geographic information can be included in the event using terms from the DwC Location class. The interacting organisms or taxa are represented by their respective instances of the DwC Occurrence and DwC Taxon classes. These classes serve as the basis of documenting data about individual organisms or species involved in the interactions. By linking these instances to an instance of Event the data schema enables the representation of a pairwise interaction. It is common that biotic interactions are sampled with a particular interest in organisms’ traits and the effects and outcomes of the interactions in which they participate. In DwC it is possible to include these data using the MeasurementOrFact class, but more complex representations can be achieved using extensions like the Extended Measurement or Fact [PAB+17] developed by Ocean Biodiversity Information System (OBIS), or the Ecological Trait-data Standard Vocabulary [SFG+19]. The eMoF is particularly useful when using the Darwin Core-Archive format because it addresses the limitations of the star-schema in representing one-to-many relationships. Instances of MeasurementOrFact class, or its extensions, can be associated with instances of the Event class to represent interaction effects and outcomes. Similarly, these measurements or facts can also be linked to instances of the Occurrence class, representing the traits of specific organisms or effects of the interaction on an individual organism or group of organisms. See Fig. 2 for a graphical representation of the data schema.
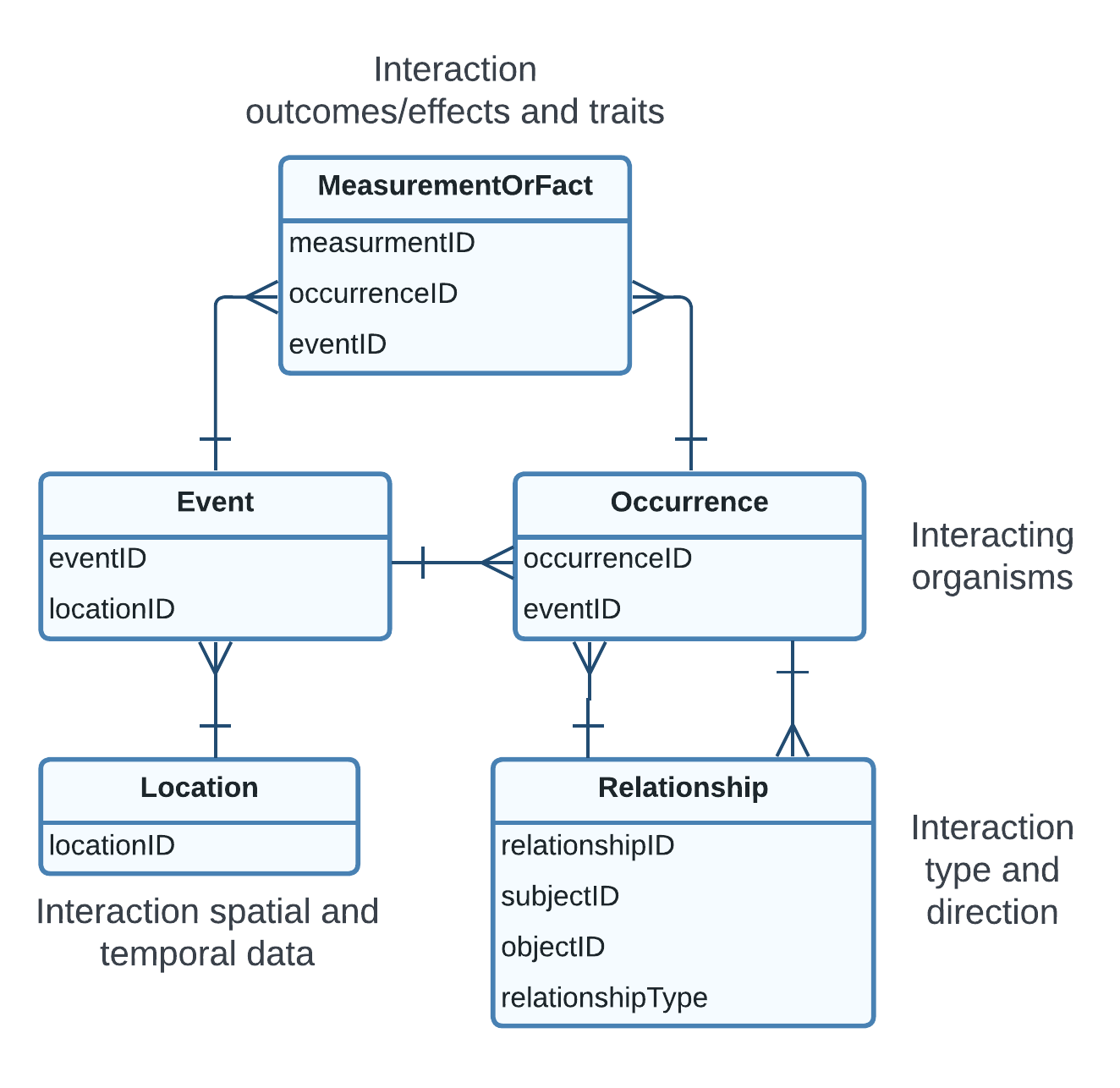
Fig. 2 Data schema for representing biotic interactions using Darwin Core. Only the identifiers and relationshipType are shown.#
The Plant-Pollinator Interactions Vocabulary (PPI)#
The Darwin Core standard does not aim to cover all use cases and specificities of biodiversity data. However, it can be used with other vocabularies to enrich data annotation and standardization of data elements which do not have corresponding concepts defined by the DwC.
However, it is common that organisms’ traits and interaction outcomes and effects data to be present in biotic interaction datasets. In addition, these data elements should be also standardized and it can be achieved by the adoption of community-specific vocabularies. In this case the REBIPP Plant-Pollinator Interaction Vocabulary (PPI) provides additional terms to the standardization of plant-animal interactions datasets.
The PPI vocabulary is recommended to be used within the Darwin Core MeasurementOrFact class as controlled vocabulary for the term measurementType. The PPI vocabulary also defines controlled vocabularies for some of its terms, and then, the terms in the controlled vocabularies should be used as values for the measurementValue term of the DwC MeasurementOrFact class.
Other vocabularies, thesauri and ontologies#
Tools#
Data standardization can be a complex and time consuming process, for that reason many tools have been created to facilitate this task. Bellow is a list a some tools which can help with data standardization:
REBIPP Plant-Pollinator Interactions Dataset Template: a Google Sheet template with controlled vocabularies for terms from DwC and PPI. In the template each row represents an interaction between a plant and an animal. Columns from the spreadsheet can be removed if they are not needed, but the inclusion of new columns is NOT RECOMMENDED.
GloBI dataset template: a simplified dataset template to make data available through Global Biotic Interactions.
Attention
REBIPP and GloBI template only simplify the process of data standardization. However, data transformation from original format to one of these templates IS NOT SUFFICIENT to consider data as standardized. The templates act as intermediate representation between original data and the standardized data which REBIPP Database and GloBI will generate when publishing the datasets. We call these template as PUBLISHING MODELS, which differ from the DATA MODELS as shown in Fig. 2
References#
- BC13
Bradley J. Butterfield and Ragan M. Callaway. A functional comparative approach to facilitation and its context dependence. Functional Ecology, 27(4):907–917, 2013. URL: doi:10.1111/1365-2435.12019 (visited on 2023-04-11).
- CBR14
Scott A. Chamberlain, Judith L. Bronstein, and Jennifer A. Rudgers. How context dependent are species interactions? Ecology Letters, 17(7):881–890, 2014. URL: doi:10.1111/ele.12279 (visited on 2023-04-11).
- Fre17
Megan E. Frederickson. Mutualisms Are Not on the Verge of Breakdown. Trends in Ecology & Evolution, 32(10):727–734, 2017. URL: doi:10.1016/j.tree.2017.07.001 (visited on 2023-04-11).
- GomezIT23
José María Gómez, José María Iriondo, and Pedro Torres. Modeling the continua in the outcomes of biotic interactions. Ecology, 104(4):e3995, 2023. URL: doi:10.1002/ecy.3995 (visited on 2023-04-11).
- Has49
Edward F Haskell. A clarification of social science. Main currents in modern thought, 7(2):45–51, 1949.
- HB15
Jason D. Hoeksema and Emilio M. Bruna. Context-dependent outcomes of mutualistic interactions. In Judith L. Bronstein, editor, Mutualism, pages 0. Oxford University Press, United Kingdom, 2015. URL: doi:10.1093/acprof:oso/9780199675654.003.0010 (visited on 2023-04-11).
- HCG+10
Jason D. Hoeksema, V. Bala Chaudhary, Catherine A. Gehring, Nancy Collins Johnson, Justine Karst, Roger T. Koide, Anne Pringle, Catherine Zabinski, James D. Bever, John C. Moore, Gail W. T. Wilson, John N. Klironomos, and James Umbanhowar. A meta-analysis of context-dependency in plant response to inoculation with mycorrhizal fungi. Ecology Letters, 13(3):394–407, 2010. URL: doi:10.1111/j.1461-0248.2009.01430.x (visited on 2023-04-11).
- Lid79
William Z. Lidicker. A Clarification of Interactions in Ecological Systems. BioScience, 29(8):475–477, 1979. URL: doi:10.2307/1307540.
- MBA14
John L. Maron, Kathryn C. Baer, and Amy L. Angert. Disentangling the drivers of context-dependent plant–animal interactions. Journal of Ecology, 102(6):1485–1496, 2014. URL: doi:10.1111/1365-2745.12305 (visited on 2023-04-11).
- PAB+17
Daphnis De Pooter, Ward Appeltans, Nicolas Bailly, Sky Bristol, Klaas Deneudt, Menashè Eliezer, Ei Fujioka, Alessandra Giorgetti, Philip Goldstein, Mirtha Lewis, Marina Lipizer, Kevin Mackay, Maria Marin, Gwenaëlle Moncoiffé, Stamatina Nikolopoulou, Pieter Provoost, Shannon Rauch, Andres Roubicek, Carlos Torres, Anton van de Putte, Leen Vandepitte, Bart Vanhoorne, Matteo Vinci, Nina Wambiji, David Watts, Eduardo Klein Salas, and Francisco Hernandez. Toward a new data standard for combined marine biological and environmental datasets - expanding OBIS beyond species occurrences. Biodiversity Data Journal, 5:e10989, 2017. URL: doi:doi:10.3897/BDJ.5.e10989 (visited on 2023-05-15).
- SSZ+22
José A Salim, Antonio M Saraiva, Paula F Zermoglio, Kayna Agostini, Marina Wolowski, Debora P Drucker, Filipi M Soares, Pedro J Bergamo, Isabela G Varassin, Leandro Freitas, Márcia M Maués, Andre R Rech, Allan K Veiga, Andre L Acosta, Andréa C Araujo, Anselmo Nogueira, Betina Blochtein, Breno M Freitas, Bruno C Albertini, Camila Maia-Silva, Carlos E P Nunes, Carmen S S Pires, Charles F dos Santos, Elisa P Queiroz, Etienne A Cartolano, Favízia F de Oliveira, Felipe W Amorim, Francisco E Fontúrbel, Gleycon V da Silva, Hélder Consolaro, Isabel Alves-dos-Santos, Isabel C Machado, Juliana S Silva, Kátia P Aleixo, Luísa G Carvalheiro, Márcia A Rocca, Mardiore Pinheiro, Michael Hrncir, Nathália S Streher, Patricia A Ferreira, Patricia M C de Albuquerque, Pietro K Maruyama, Rafael C Borges, Tereza C Giannini, and Vinícius L G Brito. Data standardization of plant–pollinator interactions. GigaScience, 11:giac043, 05 2022. URL: https://doi.org/10.1093/gigascience/giac043, arXiv:https://academic.oup.com/gigascience/article-pdf/doi/10.1093/gigascience/giac043/47161395/giac043.pdf, doi:10.1093/gigascience/giac043.
- SFG+19
Florian D. Schneider, David Fichtmueller, Martin M. Gossner, Anton Güntsch, Malte Jochum, Birgitta König-Ries, Gaëtane Le Provost, Peter Manning, Andreas Ostrowski, Caterina Penone, and Nadja K. Simons. Towards an ecological trait-data standard. Methods in Ecology and Evolution, 10(12):2006–2019, 2019. URL: doi:doi:10.1111/2041-210X.13288.
